Using AI identifies CT Lesions in less than 2 seconds: Taiwan Tech Team wins the third place in medical competition.
Professor Ching-Wei Wang's team from the Graduate Institute of Biomedical Engineering at Taiwan Tech developed a “Universal 3D Lesion Segmentation AI Model” that can be used in computed tomography (CT) scan images to quickly and accurately identify and segment multiple types of thoracoabdominal lesions. They won third place out of 632 participants in the 2024 international medical 3D CT Image AI Competition (The Universal Lesion Segmentation ’23 Challenge, ULS23).

Professor Ching-Wei Wang (right) and his team from the Graduate Institute of Biomedical Engineering at Taiwan Tech won 3rd place in this year's International Medical 3D CT Image AI Competition. On the left is Professor Wang’s student, Ding-Sheng Su, a second-year master's student at the Graduate Institute of Biomedical Engineering.
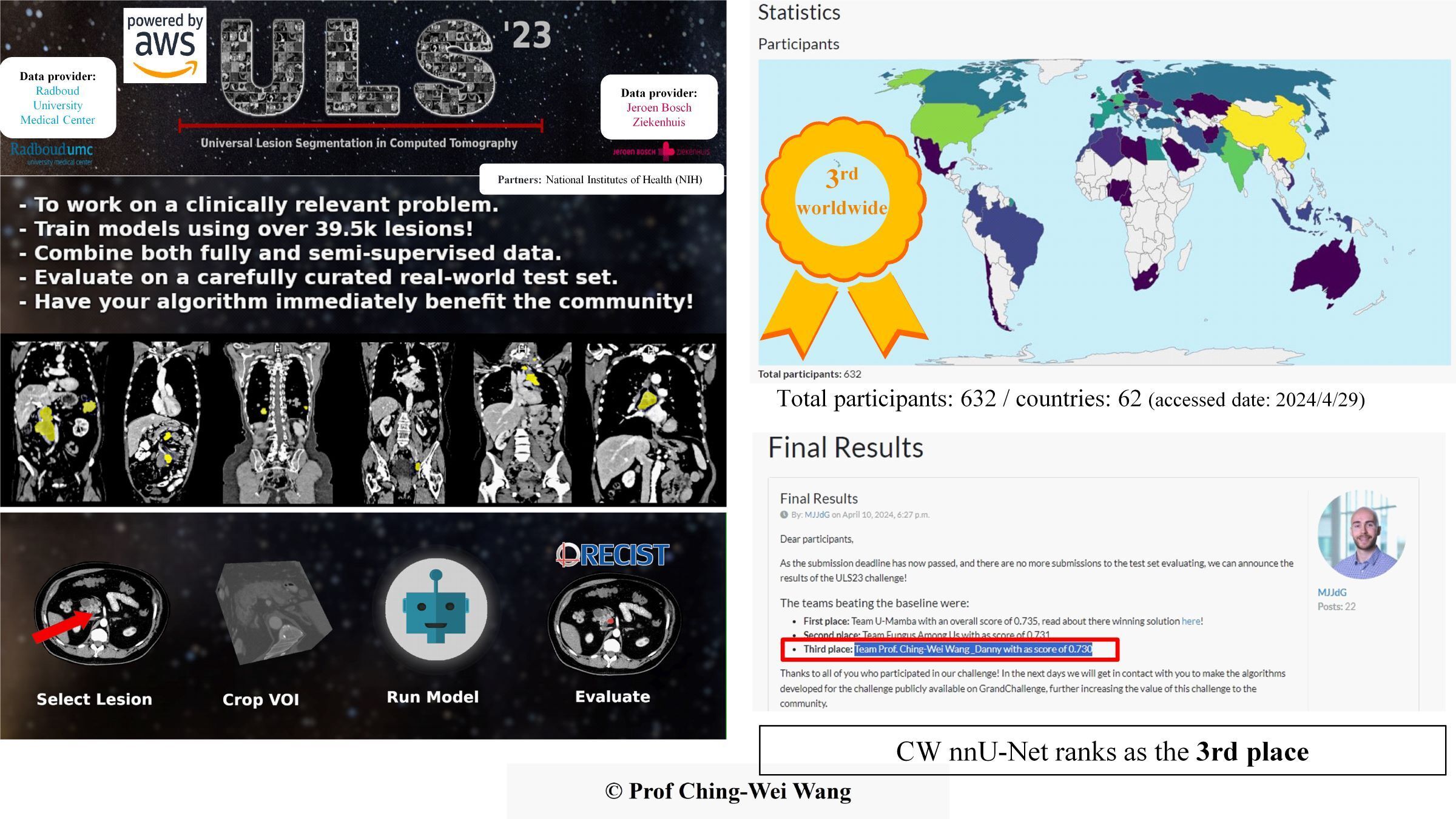
Professor Ching-Wei Wang’s team from the Graduate Institute of Biomedical Engineering at Taiwan Tech won the 3rd Place in ULS23 Competition.
Traditional CT scans face several challenges in lesion segmentation, including difficulty in lesion identification, the need for extensive expert analysis, and manual labeling of lesions, which are time-consuming and increase diagnostic inefficiency and medical costs. Manual annotation can make it easy to miss judgments due to fatigue, limited diagnostic time, and lack of experience.
Professor Ching-Wei Wang, one of the “Top 2% Global Scientists”, stated that his team has developed a “Universal 3D Lesion Segmentation AI Model” capable of accurately identifying multiple types of lesions in the chest and abdomen (including bones, pancreas, kidneys, liver, pulmonary nodules, lungs, colon, lymph nodes, and mediastinum). This model is designed for chest and abdominal CT images, automating precise 3D lesion annotation and assisting radiologists with 3D lesion marking, thus solving the problem of manual labeling that consumes a lot of labor costs.
The “Universal 3D Lesion Segmentation AI Model” not only excels in accuracy but also meets clinical efficiency needs. While traditional manual annotation takes about 30 to 60 minutes per case, the team’s AI technology processes each 3D lesion dataset in just 3.25 seconds on a Grand Challenge platform server with a single T4 GPU. It takes less than 2 seconds using a native PC with an RTX4080.
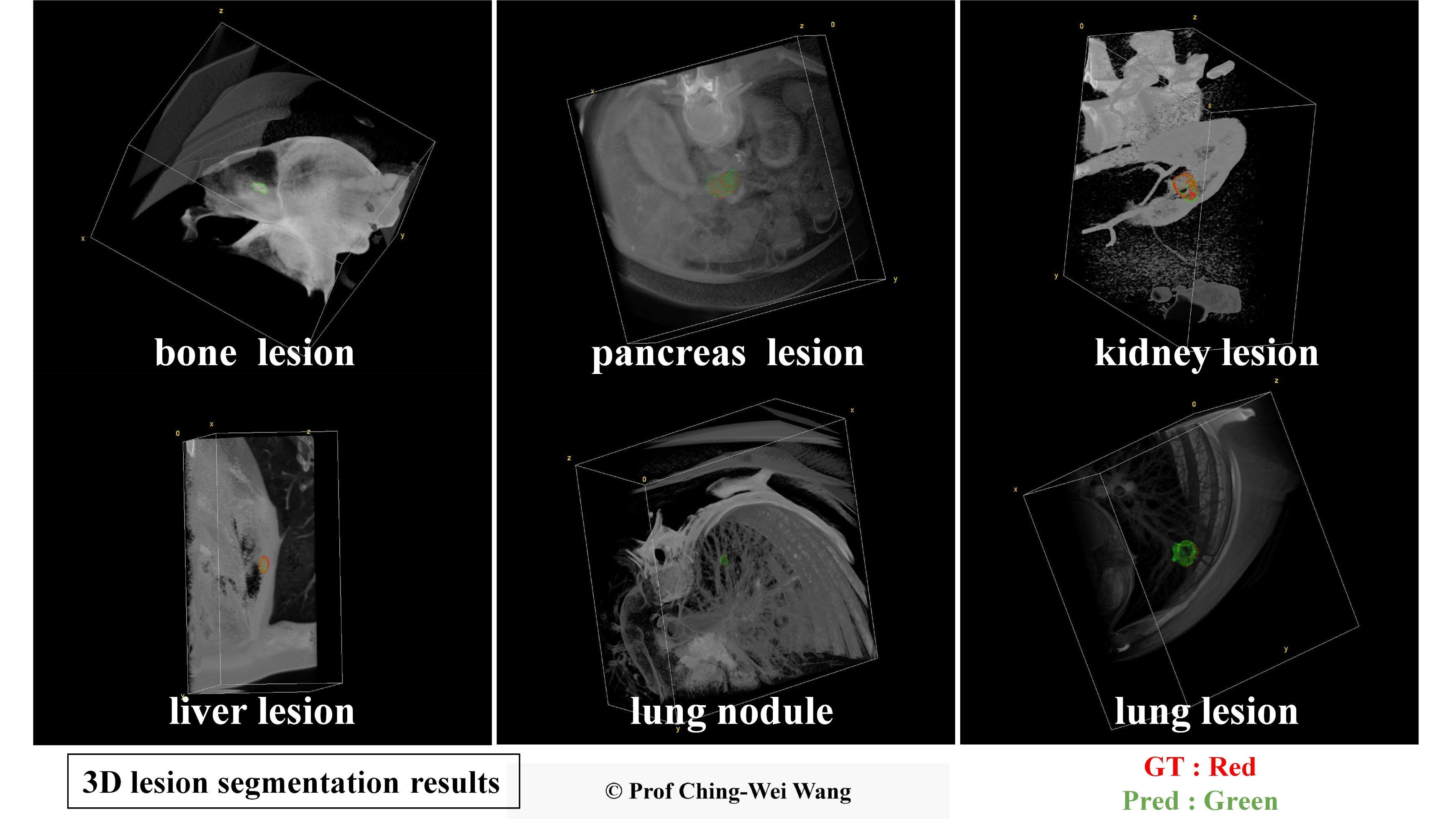
Professor Ching-Wei Wang's team has developed a universal AI model for precise 3D segmentation of various lesion types. In the images, the red contours represent the baseline truth, while the green contours represent the model's predictions.
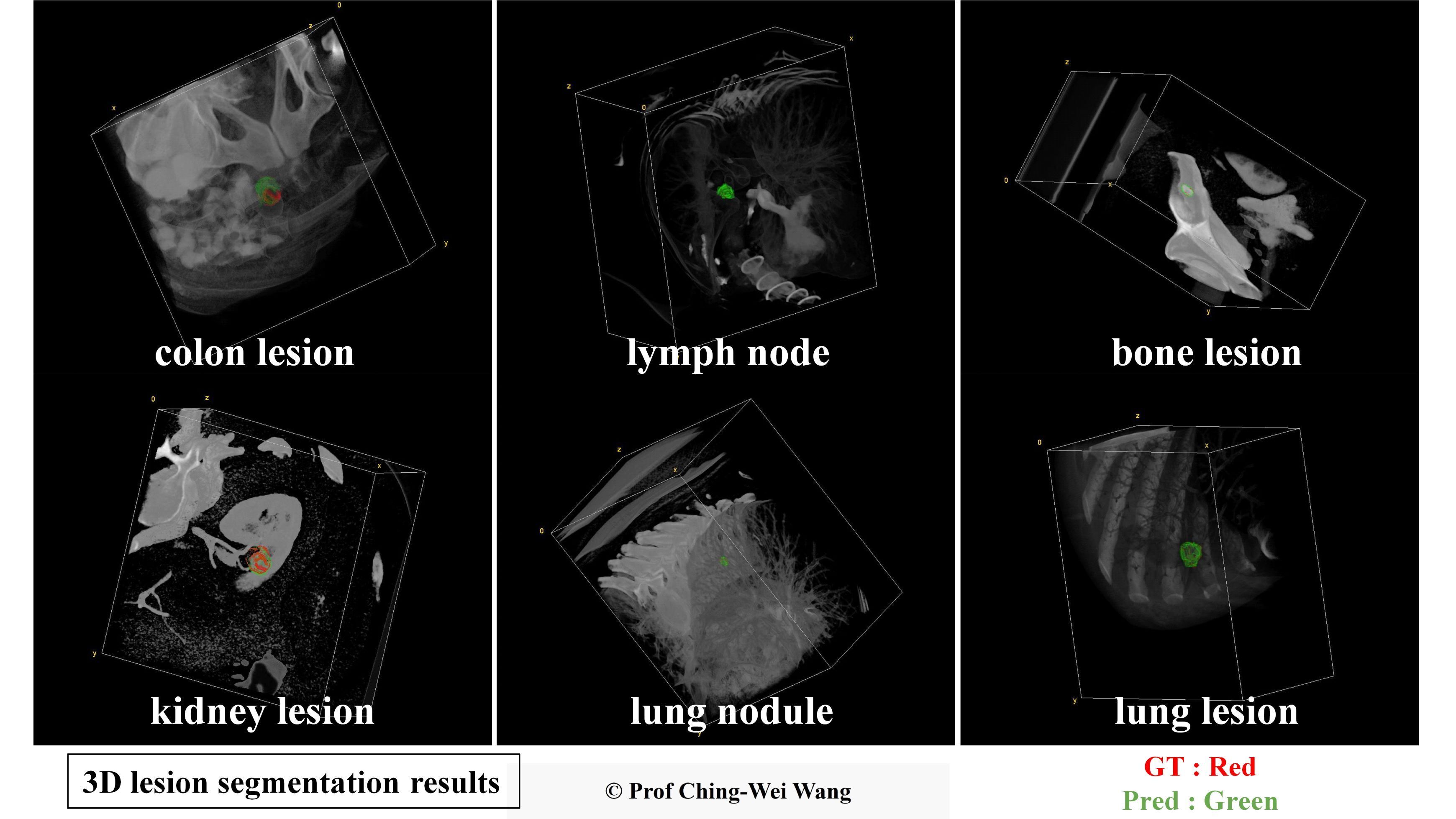
Professor Ching-Wei Wang's team has developed a universal AI model for precise 3D segmentation of various lesion types. In the images, the red contours represent the baseline truth, while the green contours represent the model's predictions.
Professor Ching-Wei Wang explained that 3D CT image lesion segmentation provides more information that helps doctors monitor lesion growth compared to 2D images, such as lesion volume, shape, and spatial location. Automated AI lesion segmentation in CT scans offers advantages over manual methods, including improved efficiency, reproducibility, accuracy, and standardization, enabling more precise quantitative analysis and facilitating the translation of research into clinical practice.
By participating in the competition, Professor Wang’s team not only enhanced their technical skills but also gained valuable experience in handling large-scale, multi-category CT image data. This has deepened their understanding of how to improve the practicality and robustness of AI models in real-world clinical applications, laying a solid foundation for future research and applications.
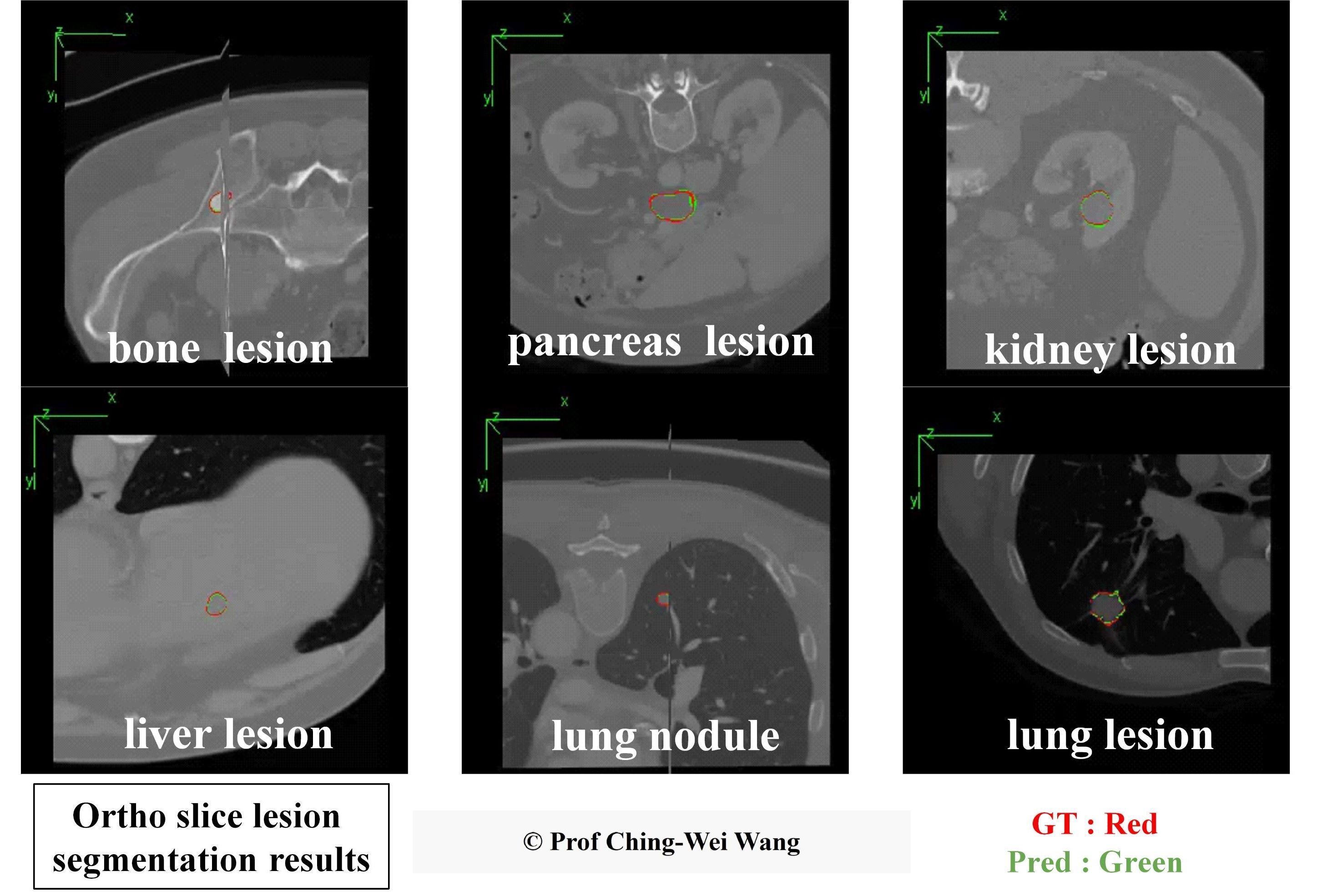
Professor Ching-Wei Wang's team from the Graduate Institute of Biomedical Engineering at Taiwan Tech has developed a universal AI model for precise 3D segmentation of various lesions. In the images, red contours represent the baseline truth, while green contours show the model's predictions.
The ULS23 competition is an international competition aimed at advancing research on universal lesion segmentation models for 3D CT imaging and is conducted on the Grand Challenge platform. The competition provides a clinical test set of 39,500 3D CT lesions, allowing participants to establish and verify multi-category universal lesion models of chest and abdominal categories, including bones, pancreas, kidneys, liver, pulmonary nodules, lungs, colon, lymph nodes, and mediastinum. The competition evaluates models based on clinical needs through multiple metrics to assess robustness and accuracy. AWS cloud computing is used for automatic and fair quantitative evaluation of participants' models using unpublished 3D CT lesion images.
Introduction video: https://www.youtube.com/watch?v=kPQLhCx9ViA
